Introduction
Biomedical data and benchmarks are highly valuable yet very limited in low-resource languages other than English, such as Vietnamese. In this work, we use a state-of-the-art translation model in English-Vietnamese to translate and produce both pretrained and supervised data in the biomedical domains. Thanks to such large-scale translation, we introduce ViPubmedT5, a pretrained EncoderDecoder Transformer model trained on 20 million translated abstracts from the high-quality public PubMed corpus. ViPubMedT5 demonstrates state-of-the-art results on two different biomedical benchmarks in summarization and acronym disambiguation. Further, we release ViMedNLI - a new NLP task in Vietnamese translated from MedNLI using the recently public En-vi translation model and carefully refined by human experts, with evaluations of existing methods against ViPubmedT5.
Pubmed and English Biomedical NLP
The Pubmed provides access to the MEDLINE database which contains titles, abstracts, and metadata from medical literature since the 1970s. The dataset consists of more than 34 million biomedical abstracts from the literature collected from sources such as life science publications, medical journals, and published online e-books. This dataset is maintained and updated yearly to include more up-to-date biomedical documents.
Pubmed Abstract has been the main dataset for almost any state-of-the-art biomedical domain specific pretrained models, for example:
ViPubmed
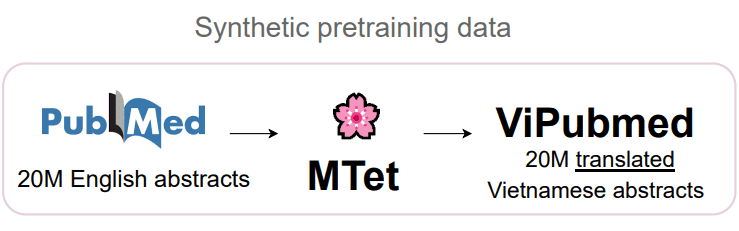
To generate a large-scale synthetic translated Vietnamese biomedical corpus, we first look into improving the existing English-Vietnamese translation system in the biomedical translation domain. After utilizing existing state-of-the-art English-Vietnamese translation work and self-training technique, we large scale translate the 20M English biomedical abstracts to Vietnamese using 4 TPUv2-8 and 4 TPUv3-8.
ViPubmedT5
With an unlabeled synthetic translated ViPubmed Corpus, we pretrain and finetune a Transformer-based language model to verify the effectiveness of our approach in enriching Vietnamese biomedical domain with translation data.
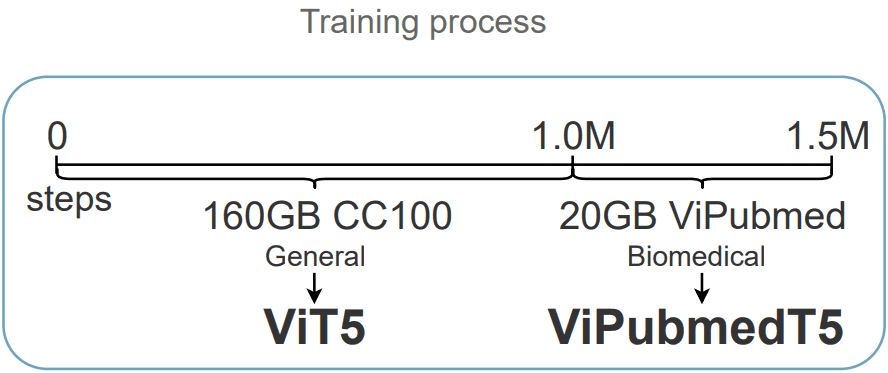
We initialize the checkpoint and architecture from ViT5, a Vietnamese T5 model. Then, we pretrain the model on the synthetic translated ViPubmed corpus for 500k steps. We train ViPubmedT5 using the spans masking learning objective. During self-supervised training, spans of biomedical text sequences are randomly masked (with sentinel tokens). The target sequence is formed as the concatenation of the same sentinel tokens and the real masked spans/tokens.
Finetunning Results
We finetune and benchmark our pretrained ViPubmedT5 model on two public Vietnamese biomedical-domain datasets acrDrAid & FAQSum (from ViHealthBERT work) and our released ViMedNLI dataset.
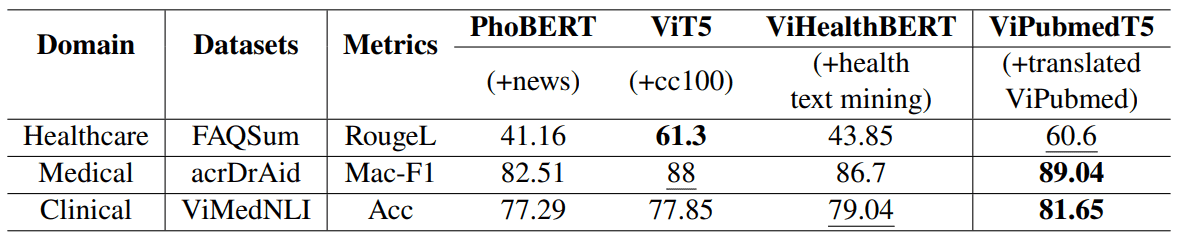 Notes: The best scores are in bold, and the second best scores are underlined. PhoBERT
& ViHealthBERT scores on FAQSum and acrDrAid are from ViHealthBERT paper
Notes: The best scores are in bold, and the second best scores are underlined. PhoBERT
& ViHealthBERT scores on FAQSum and acrDrAid are from ViHealthBERT paper
The main takeaway is that training on synthetic translated biomedical data allows ViPubmedT5 to learn a better biomedical context representation. As a result, ViPubmedT5 achieves state-of-the-art in Medical and Clinical contexts while performing slightly worse than ViT5 in healthcare topics.
For both medical and clinical datasets, ViPubmedT5 outperforms other existing models. There are also notable improvements from the general domain ViT5 to ViPubmedT5 (88->89.04 in acrDrAid and 77.85->81.65 in ViMedNLI). This indicates that the translated ViPubmed corpus contains biomedical knowledge that low-resource Vietnamese pretrained models can leverage.
Scaling to Other Languages
Our novel way of utilizing a state-of-the-art NMT system to generate synthetic translated medical data for pretrained models is not limited to the Vietnamese language and is scalable to many other low-resource languages. Recent works focusing on improving the quality of multiple low-resource NMT systems (M2M100 or NLLB) make the approach discussed in this paper more practical to produce synthetic translated biomedical data and enrich the Biomedical NLP research knowledge in multiple low-resource languages.
Acknowledgements
We would like to thank the Google TPU Research Cloud (TRC) program and Soonson Kwon (Google ML Ecosystem programs Lead) for their supports.
Refer to this work
@misc{vipubmed,doi = {10.48550/ARXIV.2210.05598},url = {https://arxiv.org/abs/2210.05598},author = {Phan, Long and Dang, Tai and Tran, Hieu and Trinh, Trieu H. and Phan, Vy and Chau, Lam D. and Luong, Minh-Thang},keywords = {Computation and Language (cs.CL), Artificial Intelligence (cs.AI), FOS: Computer and information sciences, FOS: Computer and information sciences},title = {Enriching Biomedical Knowledge for Low-resource Language Through Large-Scale Translation},publisher = {arXiv},year = {2022},copyright = {Creative Commons Attribution 4.0 International}}